CURRENT PROJECTS
Two-photon Collinearly Multiplexed Beams (2pCOMB)
Reference: "Bridging scales in scattering tissues via multifocal two-photon microscopy", bioRxiv preprint (2020)
Imaging biological systems at subcellular resolution and across scales is essential to understanding how cells form tissues, organs, and organisms. However, existing large-scale optical techniques often require harsh tissue-clearing methods that cause significant morphological changes, compromise the integrity of cell membranes, and reduce the signal of fluorescent proteins. We demonstrated multifocal two-photon microscopy that enabled imaging mesoscopic scattering samples in their native tissue environment at high resolution and high speed.
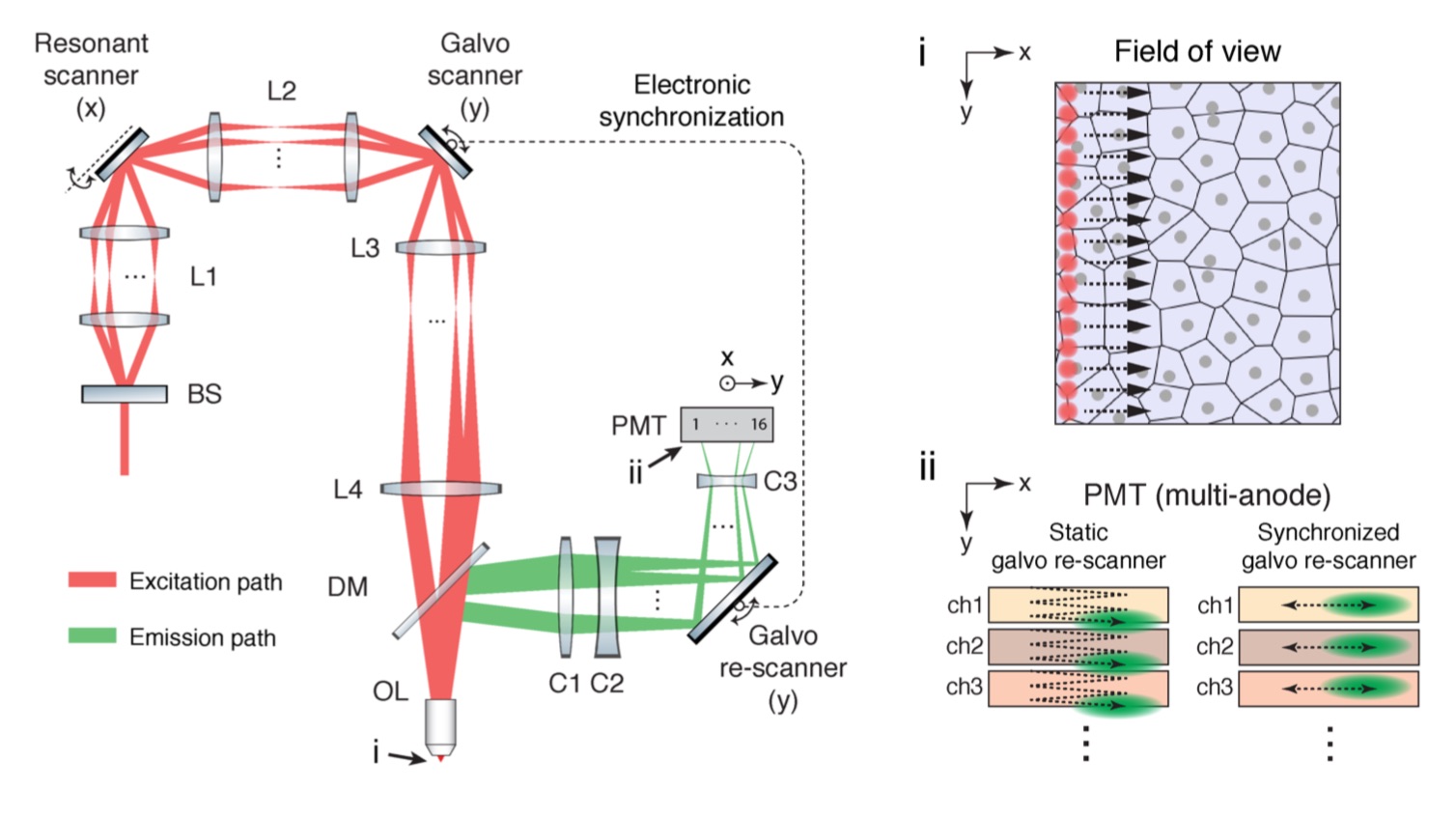 Simplified optical layout of two-photon microscopy with collinearly multiplexed beams (2pCOMB). A beam-splitter (BS) creates a linear array of 16 laser beams. The beams pass through relay lenses (L1–L4), focus onto the sample through an objective lens (OL) and scan the field of view with a ‘laser comb’ of 16 excitation foci (i). This beam configuration allows a 16-fold increase in throughput compared with a conventional single-beam scheme. The 16 fluorescence signals return through OL, reflect off a dichroic mirror (DM), and focus onto a 16-anode PMT through collector lenses (C1–C3). A galvanometer mirror in the y-axis re-scans the fluorescence emission to minimize the crosstalk between adjacent anodes of the PMT (ii)
Simplified optical layout of two-photon microscopy with collinearly multiplexed beams (2pCOMB). A beam-splitter (BS) creates a linear array of 16 laser beams. The beams pass through relay lenses (L1–L4), focus onto the sample through an objective lens (OL) and scan the field of view with a ‘laser comb’ of 16 excitation foci (i). This beam configuration allows a 16-fold increase in throughput compared with a conventional single-beam scheme. The 16 fluorescence signals return through OL, reflect off a dichroic mirror (DM), and focus onto a 16-anode PMT through collector lenses (C1–C3). A galvanometer mirror in the y-axis re-scans the fluorescence emission to minimize the crosstalk between adjacent anodes of the PMT (ii)
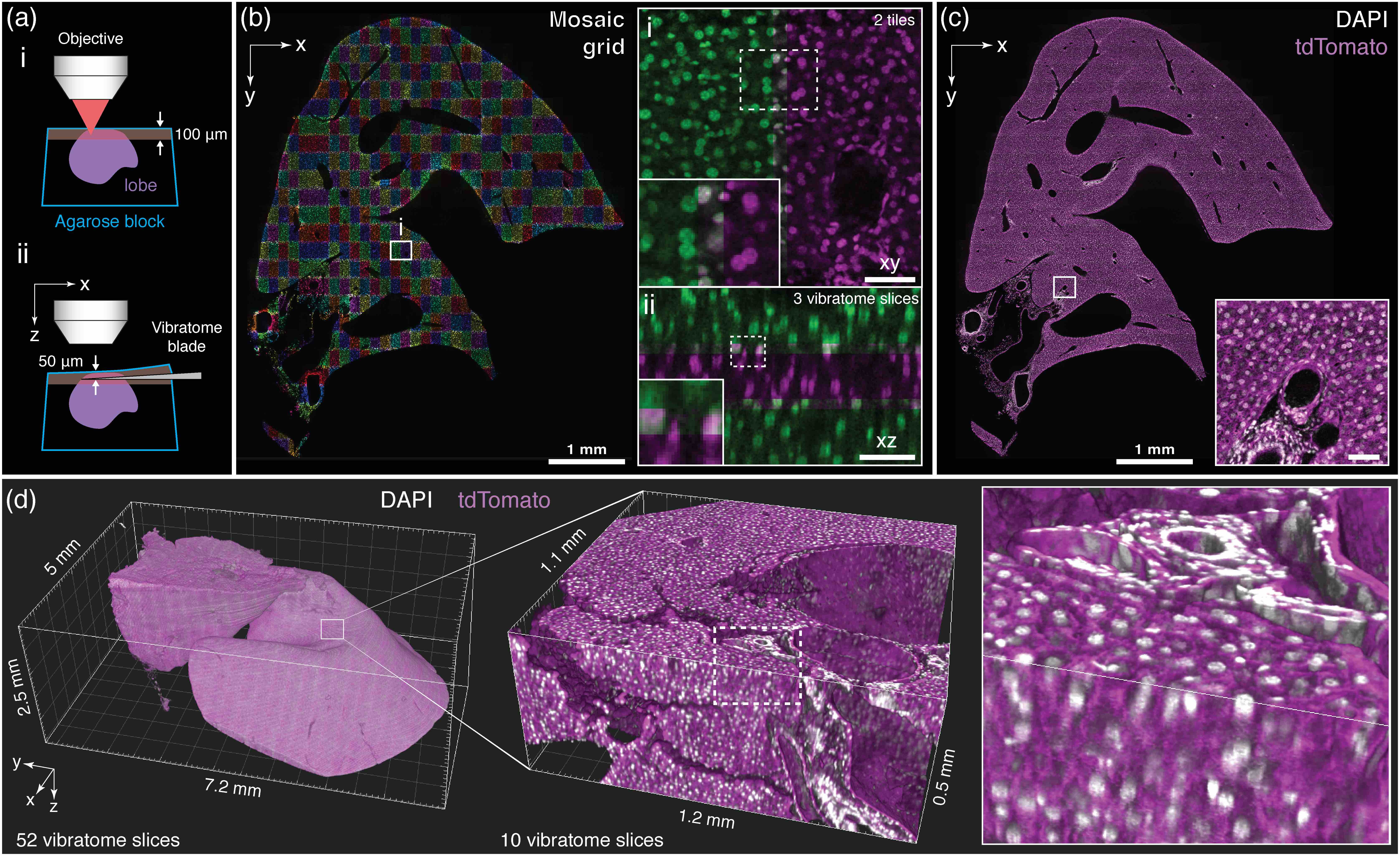 Multiscale imaging and digital reconstruction of a mouse liver lobe. a, An entire liver lobe was imaged iteratively by scanning its surface at high speed via 2pCOMB and then physically removing the tissue through in situ tissue sectioning (i and ii, respectively). b, Cell nuclei across a full xy-section of the lobe acquired via mosaic imaging with a z-stack of 150 μm × 280 μm × 100 μm in xyz. The enlarged views show co-aligned z-stacks in xy (i) and xz (ii) with an overlap of 10%, 15%, and 17% in xyz. Co-localized nuclei between adjacent stacks appear in grey. c, Cell nuclei (grey) across the same plane as in b with the stacks fused and overlaid with a second fluorescent channel showing the plasma membranes (magenta). The cross-section is located at approximately the center of the lobe. d, 3D rendering of the entire liver lobe comprised of 52 vibratome slices. The sample was imaged with a voxel size of 0.5 μm × 0.5 μm × 1 μm throughout the full volume. The lobe has been downsampled for display purposes (2 μm/voxel for the whole lobe and 1 μm/voxel for the enlarged volume). Scale bars, 50 μm, unless otherwise stated.
Multiscale imaging and digital reconstruction of a mouse liver lobe. a, An entire liver lobe was imaged iteratively by scanning its surface at high speed via 2pCOMB and then physically removing the tissue through in situ tissue sectioning (i and ii, respectively). b, Cell nuclei across a full xy-section of the lobe acquired via mosaic imaging with a z-stack of 150 μm × 280 μm × 100 μm in xyz. The enlarged views show co-aligned z-stacks in xy (i) and xz (ii) with an overlap of 10%, 15%, and 17% in xyz. Co-localized nuclei between adjacent stacks appear in grey. c, Cell nuclei (grey) across the same plane as in b with the stacks fused and overlaid with a second fluorescent channel showing the plasma membranes (magenta). The cross-section is located at approximately the center of the lobe. d, 3D rendering of the entire liver lobe comprised of 52 vibratome slices. The sample was imaged with a voxel size of 0.5 μm × 0.5 μm × 1 μm throughout the full volume. The lobe has been downsampled for display purposes (2 μm/voxel for the whole lobe and 1 μm/voxel for the enlarged volume). Scale bars, 50 μm, unless otherwise stated.
Here are some
photos of the lab and microscope.
PAST PROJECTS
During my Ph.D. studies, I investigated emergent phenomena in ultracold quantum gases. I used advanced electro-optical and magneto-optical techniques to trap, cool, and manipulate ultracold matter with nano-scale precision (both, spatial and temporal)

For my B.S. dissertation, I used scanning tunneling microscopy to measure, analyze, and model the surface of thin conductive films to investigate size effects on nanomaterials.




